Standing guard between a cell's nucleus and its main chamber, called the cytoplasm, are thousands of behemoth protein structures called nuclear pore complexes, or NPCs. NPCs are like the bouncers of a cell's nucleus, tightly guarding exactly what goes in and out. Each structure contains about 1,000 protein molecules, making NPCs some of the biggest protein complexes in our bodies. One of the most notable clients of NPCs is a class of molecules known as messenger RNAs, or mRNAs. These are the messengers that carry genetic instructions from the nucleus to the cytoplasm, where they are then translated into proteins.
But how the NPC transports the mRNAs out of the nucleus is still a mystery.
"The mRNAs are one of the largest cargoes carried through NPCs, and the whole process occurs in just a fraction of a second," says André Hoelz, professor of chemistry at Caltech, a Heritage Medical Research Institute (HMRI) Investigator, and a Howard Hughes Medical Institute (HHMI) Faculty Scholar. "How this works has been one of the greatest unsolved problems in biology."
NPCs are associated with several diseases. Mutations to proteins within the complex have been linked to motor neuron diseases such as amyotrophic lateral sclerosis (ALS), and people with Huntington's disease are known to have defects in the function of their NPCs.

In a new study in the June 13 issue of Nature Communications, Hoelz and his group—spearheaded by Daniel Lin (PhD '17), a former graduate student at Caltech now at Whitehead Institute for Biomedical Research at MIT, and Sarah Cai, an undergraduate student at Caltech—report the first atomic-scale look at the specific components of human NPCs responsible for dropping mRNAs off in the cytoplasm. For an mRNA to be transported through an NPC, it must be tagged with a nuclear export factor, a type of small protein. That tag is like a ticket that allows the mRNA to enter the central transport channel of the NPC. Once the mRNA reaches the cytoplasmic side, it must surrender the ticket—otherwise, the mRNA could travel back into the nucleus, and the proteins it encodes wouldn't get made.
Through a series of experiments involving X-ray crystallography, biochemistry, enzymology, and other methodologies, the researchers were able to show how this process of un-tagging the mRNA molecules works in human cells for the first time.
"It's as if we had snapshots before, and now we have a movie showing us exactly what happens at the molecular scale when mRNAs are dropped off in the cell's cytoplasm," says Lin.
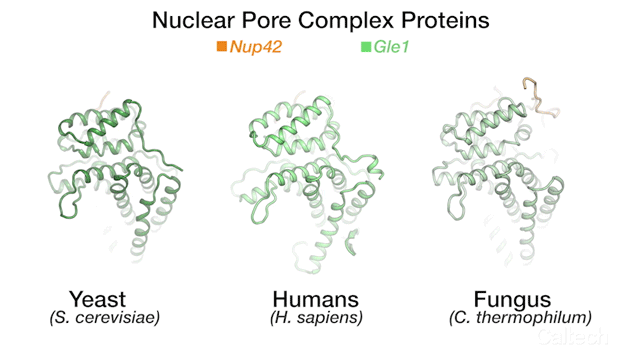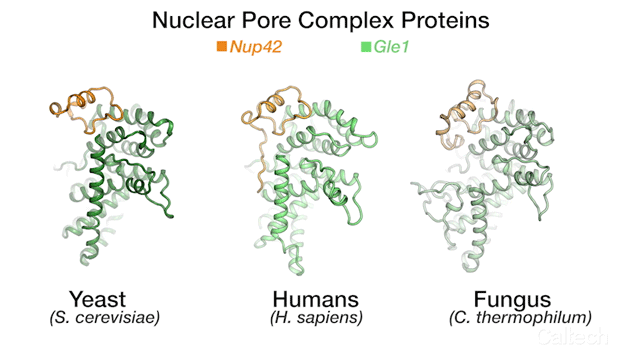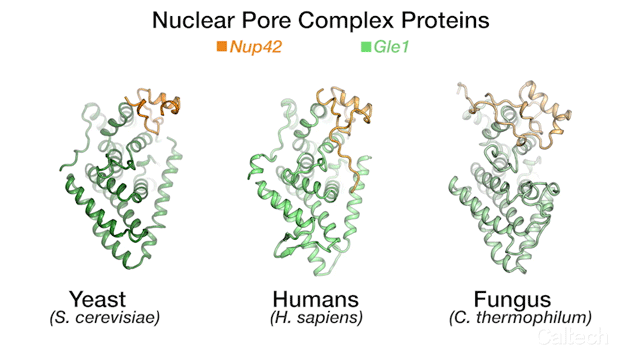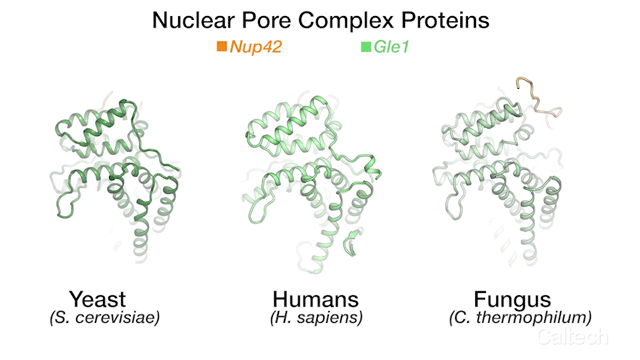
The team's new findings were made possible by obtaining a series of crystal structures of a few key protein components of a human NPC. One of those components is called Gle1. The three-dimensional structure of this protein had been obtained before in yeast, but doing so for its human variant had remained a challenge. By studying the biochemical properties of yeast Gle1, the researchers were able to figure out that another protein, called Nup42, was required to stabilize Gle1. Knowing this, the team was able to purify human Gle1 from cells in high quantities for the first time, and then, using Caltech's Molecular Observatory beamline at the Stanford Synchrotron Radiation Lightsource, obtain its crystal structure.
"Even with billions of years of evolution between yeast and humans, there are still aspects of our bio-machinery that remain the same," says Lin.
With the ability to purify human Gle1, the researchers set about studying how mutations affect its structure. They looked at several specific mutations of Gle1 known to be associated with a motor neuron disease called lethal contracture congenital syndrome 1 (LCCS1) and discovered that the mutated versions of the protein were not as stable.
"Gle1 is essential for life to function properly," says Hoelz, "so any mutations that cause it to be less stable are going to cause problems."
The researchers then looked at the structure of Gle1 bound to a protein called DDX19—which is responsible for un-tagging the mRNA molecules after they pass through the NPC. Gle1 is required to activate DDX19, and—until now—it was thought that a small molecule called inositol hexaphosphate (IP6) acted like a tether between Gle1 and DDX19, allowing the activation to occur.
"We found that IP6 was not required in humans, and that was a surprise because it is required in yeast, and IP6 dependence was previously believed to occur across all species," says Cai. "While there are some similarities between yeast and human proteins, there are also crucial differences."
What's more, the new research shows in atomic-level detail exactly how the un-tagging of the mRNA works. This kind of structural information could be used in the future to help in the design of therapeutic drugs for motor neuron diseases.
Hoelz says that Lin and Cai really exceeded expectations for this research. "They wanted to discover something new, and they went above and beyond with this project," he says. "They made it happen. This is a Caltech moment."
Other co-authors of the Nature Communications paper, titled "Structural and functional analysis of mRNA export regulation by the nuclear pore complex," include Ana Correia, formerly a postdoctoral scholar at Caltech now working at Amgen, and Caltech graduate students Ferdinand Huber and Claudia Jette. Funding comes from the National Institutes of Health, Amgen, a Margaret Leighton Summer Undergraduate Research Fellowship (SURF), a John Stauffer SURF, and a Camille Dreyfus Teacher-Scholar Award.
 The three-dimensional structures for three proteins in the human nuclear pore complex: Gle1 (green), Nup42 (yellow), and DDX19 (pink).
Credit: Hoelz Lab/Dan Lin/Caltech
A movie showing the three-dimensional structures, and interactions, for three proteins in the human nuclear pore complex—a gatekeeper in the cell that ushers mRNA molecules from the nucleus to the cytoplasm. Gle1, which is stabilized by Nup42, helps activate DDX19, priming it to bind mRNA molecules. Ultimately, DDX19 helps to send mRNAs along their way, so that they can be translated into proteins in the cytoplasm.
Credit: Hoelz Lab/Dan Lin/Caltech
The three-dimensional structures for three proteins in the human nuclear pore complex: Gle1 (green), Nup42 (yellow), and DDX19 (pink).
Credit: Hoelz Lab/Dan Lin/Caltech
A movie showing the three-dimensional structures, and interactions, for three proteins in the human nuclear pore complex—a gatekeeper in the cell that ushers mRNA molecules from the nucleus to the cytoplasm. Gle1, which is stabilized by Nup42, helps activate DDX19, priming it to bind mRNA molecules. Ultimately, DDX19 helps to send mRNAs along their way, so that they can be translated into proteins in the cytoplasm.
Credit: Hoelz Lab/Dan Lin/Caltech

