Proof-of-concept project paves the way for the integration of molecules with computer chips
Engineers have developed a technique that allows them to precisely place microscopic devices formed from folded DNA molecules in not only a specific location but also in a specific orientation.
As a proof-of-concept, they arranged more than 3,000 glowing moon-shaped nanoscale molecular devices into a flower-shaped instrument for indicating the polarization of light. Each of 12 petals pointed in a different direction around the center of the flower, and within in each petal about 250 moons were aligned to the direction of the petal. Because each moon only glows when struck by polarized light matching its orientation, the end result is a flower whose petals light up in sequence as the polarization of light shined upon it is rotated. The flower, which spans a distance smaller than the width of a human hair, demonstrates that thousands of molecules can be reliably oriented on the surface of a chip.
This method for precisely placing and orienting DNA-based molecular devices may make it possible to use these molecular devices to power new kinds of chips that integrate molecular biosensors with optics and electronics for applications such as DNA sequencing or measuring the concentrations of thousands of proteins at once.
The research, published on February 19 by the journal Science, builds on more than 15 years of work by Caltech's Paul Rothemund (BS '94), research professor of bioengineering, computing and mathematical sciences, and computation and neural systems, and his colleagues. In 2006, Rothemund showed that DNA could be directed to fold itself into precise shapes through a technique dubbed DNA origami. In 2009, Rothemund and colleagues at IBM Research Almaden described a technique through which DNA origami could be positioned at precise locations on surfaces. To do so, they used a printing process based on electron beams and created "sticky" patches having the same size and shape as the origami did. In particular, they showed that origami triangles bound precisely at the location of triangular sticky patches.
Next, Rothemund and Ashwin Gopinath, formerly a Caltech senior postdoctoral scholar and now an assistant professor at MIT, refined and extended this technique to demonstrate that molecular devices constructed from DNA origami could be reliably integrated into larger optical devices. "The technological barrier has been how to reproducibly organize vast numbers of molecular devices into the right patterns on the kinds of materials used for chips," says Rothemund.
In 2016, Rothemund and Gopinath showed that triangular origami carrying fluorescent molecules could be used to reproduce a 65,000-pixel version of Vincent van Gogh's The Starry Night. In that work, triangular DNA origami were used to position fluorescent molecules within bacterium-sized optical resonators; precise placement of the fluorescent molecules was critical since a move of just 100 nanometers to the left or right would dim or brighten the pixel by more than five times.
But the technique had an Achilles' heel: "Because the triangles were equilateral and were free to rotate and flip upside-down, they could stick flat onto the triangular sticky patch on the surface in any of six different ways. This meant we couldn't use any devices that required a particular orientation to function. We were stuck with devices that would work equally well when pointed up, down, or in any direction," says Gopinath. Molecular devices intended for DNA sequencing or measuring proteins absolutely have to land right side up, so the team's older techniques would ruin 50 percent of the devices. For devices also requiring a unique rotational orientation, such as transistors, only 16 percent would function.
The first problem to solve, then, was to get the DNA origami to reliably land with the correct side facing up. "It's a bit like guaranteeing toast always magically lands butter side up when thrown on the floor," says Rothemund. To the researchers surprise, coating origami with a carpet of flexible DNA strands on one side enabled more than 95 percent of them to land face up. But the problem of controlling rotation remained. Right triangles with three different edge lengths were the researchers' first attempt at a shape that might land in the preferred rotation.
However, after wrestling to get just 40 percent of right triangles to point in the correct orientation, Gopinath recruited computer scientists Chris Thachuk of the University of Washington, co-author of the Science paper, and a former Caltech postdoc; and David Kirkpatrick of the University of British Columbia, also a co-author of the Science paper. Their job was to find a shape which would only get stuck in the intended orientation, no matter what orientation it might land in. The computer scientists' solution was a disk with an off-center hole, which the researchers termed a "small moon." Mathematical proofs suggested that, unlike a right triangle, small moons could smoothly rotate to find the best alignment with their sticky patch without getting stuck. Lab experiments verified that over 98 percent of the small moons found the correct orientation on their sticky patches.
The team then added special fluorescent molecules that jam themselves tightly into the DNA helices of the small moons, perpendicular to the axis of the helices. This ensured that the fluorescent molecules within a moon were all oriented in the same direction and would glow most brightly when stimulated with light of a particular polarization. "It's as if every molecule carries a little antenna, which can accept energy from light most efficiently only when the polarization of light matches the orientation of the antenna," says Gopinath. This simple effect is what enabled the construction of the polarization-sensitive flower.
With robust methods for controlling the up-down and rotational orientation of DNA origami, a wide range of molecular devices may now be cheaply integrated into computer chips in high yield for a variety of potential applications. For example, Rothemund and Gopinath have founded a company, Palamedrix, to commercialize the technology for building semiconductor chips that enable simultaneous study of all the proteins relevant to human health. Caltech has filed patent applications for the work.
The paper is titled "Absolute and arbitrary orientation of single-molecule shapes." Co-authors at Caltech include Harry Atwater, the Howard Hughes Professor of Applied Physics and Materials Science, and former graduate student Anna Mitskovets (PhD '20). This work was supported by the Office of Naval Research, the Air Force Office of Scientific Research, the National Science Foundation, the Orr Family Foundation, the Abedin Institute, and a Banting Postdoctoral Fellowship. Fabrication was done at the Kavli Nanoscience Institute's nanofabrication laboratory.
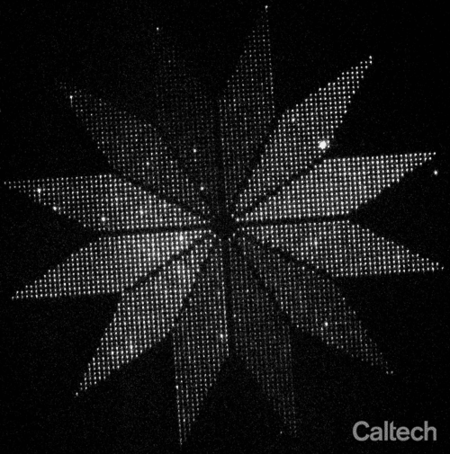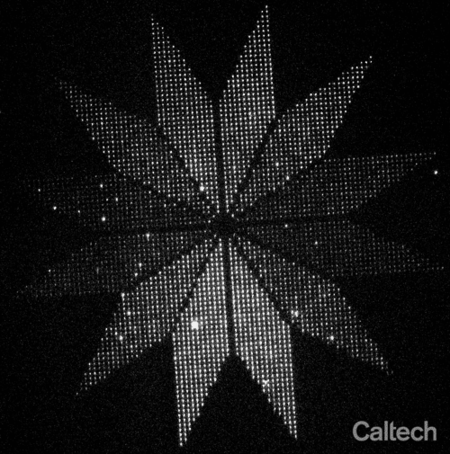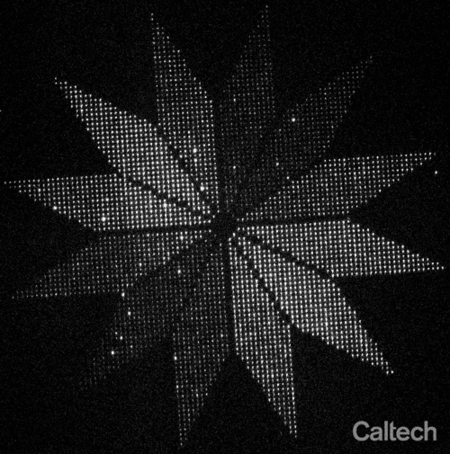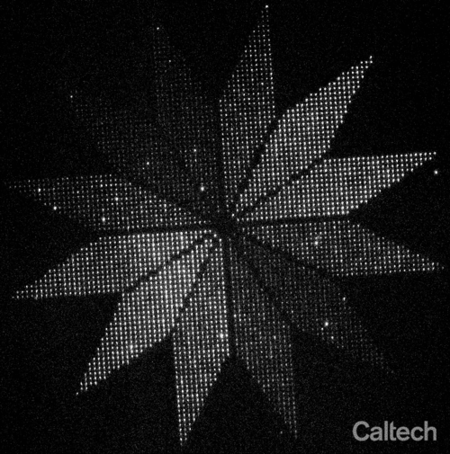 Researchers placed more than 3,000 glowing moon-shaped nanoscale molecular devices into a flower-shaped instrument for indicating the polarization of light. The "moons" in each of the 12 petals points in a different direction, and only glows when struck by polarized light matching its orientation. The end result is a flower whose petals light up in sequence as the polarization of light shined upon it is rotated. The flower, which spans a distance smaller than the width of a human hair, demonstrates that thousands of molecules can be reliably oriented on the surface of a chip.
Credit: Ashwin Gopinath/Caltech
Researchers placed more than 3,000 glowing moon-shaped nanoscale molecular devices into a flower-shaped instrument for indicating the polarization of light. The "moons" in each of the 12 petals points in a different direction, and only glows when struck by polarized light matching its orientation. The end result is a flower whose petals light up in sequence as the polarization of light shined upon it is rotated. The flower, which spans a distance smaller than the width of a human hair, demonstrates that thousands of molecules can be reliably oriented on the surface of a chip.
Credit: Ashwin Gopinath/Caltech

