Every living thing on the planet contains DNA, the molecular sequence that encodes the genetic blueprint of an organism. Genome sequencing can reveal your likelihood of getting certain diseases like Alzheimer's, and it can tell you whether you have straight or curly hair or are likely to sneeze when sunlight hits your eyes. But for all this information, scientists only understand the functions of a small portion of the genome. The bacterium Escherichia coli (E. coli) is widely considered the most well-studied organism on Earth, but still, scientists have no idea how more than half of E. coli's genes are regulated.
Now, recent research from Caltech illustrates a new technique to help crack the code of certain mysterious regions of DNA called noncoding DNA sequences. Many mutations in these poorly understood regions have been implicated in disease in organisms such as humans, so understanding the function of noncoding DNA is critical.
The work was done in the laboratory of Rob Phillips, Fred and Nancy Morris Professor of Biophysics, Biology, and Physics. A paper describing the research appears online on May 4, ahead of print in the journal Proceedings of the National Academy of Sciences.
"Humans have such a wide variety of cells—muscle cells, neurons, photoreceptors, blood cells, to name a few," says Phillips. "They all have the same DNA, so how do they each turn out so differently? The answer lies in the fact that genes can be regulated—turned on or off, dialed up and dialed down—differently in different tissues. Until now, there have been no general principles to help us understand how this regulation was encoded."
The most well-studied parts of the genome are the so-called coding regions—the genes that encode for the production of the proteins that allow a cell to function.
However, more than 50 percent of the genes in E. coli have noncoding regions whose functions remain completely mysterious. These regions of the DNA contain sites where proteins called transcription factors bind and are able to dial up or down expression of other genes—in other words, noncoding regions contain information about how the genome regulates itself.
In the new work, postdoctoral scholar Nathan Belliveau (PhD '18) applied a method called Sort-Seq to mutate small pieces of noncoding regions in E. coli and determine which regions contain binding sites. Binding sites are the locations where specialized proteins that are involved in transcription—the first step in the process of gene expression—attach to DNA.
First, the researchers cut out potentially interesting sections of noncoding DNA that they wanted to learn about. To these, they attached DNA encoding for the production of a glowing green fluorescent protein (GFP). Then, each little engineered section of DNA was placed inside an individual E. coli bacterium, causing it to produce these green proteins.
When Belliveau randomly mutated parts of the unknown regions, he noted observable changes in the amount of GFP produced in some of the bacteria, indicating that the mutated DNA is altering the level of gene expression. Through DNA sequencing, the researchers were then able to pinpoint the exact location of these important mutations and use this information to identify new binding sites.
Phillips gives a literary analogy: "This is as if I went through a book, randomly took 10 percent of the letters in words, and changed them. If the first letter of 'walk' gets changed to a T, making the word 'talk,' then you change the meaning of the word completely—your comprehension changes. We wanted to know: Which parts of the genome affect cellular comprehension the most?"
After examining many noncoding regions to determine binding sites, the team aimed to match the regions with the corresponding proteins that bind there.
"This was literally like finding a needle in a haystack," says Phillips. "There are roughly 3 million proteins in E. coli, and maybe 10 copies of a particular protein that will correspond to a given binding site. That's finding one protein in 300,000 proteins."
Belliveau developed a method to find the proverbial needle: He took a piece of noncoding DNA that contained a binding site, poured the contents of an E. coli cell over that DNA, and then identified the protein that had stuck to the site.
"This work is a demonstration that we can use our approach to go from nothing—complete ignorance—to actually understanding mechanisms of regulation," says Belliveau. "The next step is to try to scale this up to allow us to go after the entire genome."
"We live in a genomic era," says Phillips. "We have to be able to figure out how, where, and when genes are turned off and on."
The paper is titled "A systematic approach for dissecting the molecular mechanisms of transcriptional regulation in bacteria." Other Caltech co-authors are graduate students Stephanie Barnes and William Ireland, senior bioinformatician Michael Sweredoski, senior lab manager and research scientist Annie Moradian, and former Proteome Exploration Laboratory director Sonja Hess. Additional co-authors include Daniel Jones (PhD '14) of Uppsala University and Justin Kinney of Cold Spring Harbor Laboratory. Funding was provided by La Fondation Pierre-Gilles de Gennes, the Donna and Benjamin M. Rosen Bioengineering Center at Caltech, the National Institutes of Health, the Gordon and Betty Moore Foundation, Caltech's Beckman Institute, and the Howard Hughes Medical Institute.
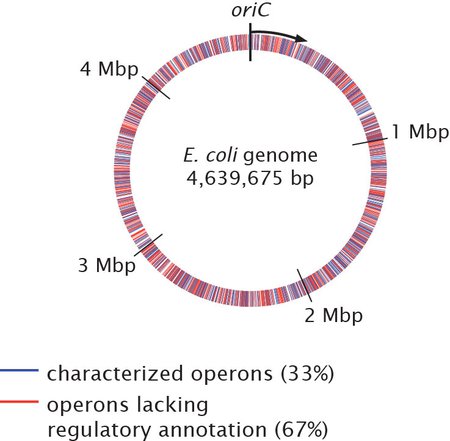 The circular E. coli genome. Sets of genes, or operons, which lack regulatory understanding are shown in red. Though E. coli is the most well-studied organism on the planet, scientists do not understand the regulatory mechanisms of 67 percent of E. coli's genes.
Credit: Courtesy of the Phillips laboratory
The circular E. coli genome. Sets of genes, or operons, which lack regulatory understanding are shown in red. Though E. coli is the most well-studied organism on the planet, scientists do not understand the regulatory mechanisms of 67 percent of E. coli's genes.
Credit: Courtesy of the Phillips laboratory

